Gupta Lab
Multi-omics data analysis and integration
Our group specializes in multi-omics data analysis and integration, with a focus on interpretable machine learning methods.
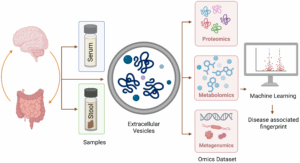
We are contributing to the translational project “Tracking Multiple Sclerosis in the Gut: Unraveling Disease-Associated Patterns in the Fecal Vesiculome,” which Teresa Bodenmeier is carrying out in collaboration with Prof. Claudia Günther and Prof. Veit Rothhammer , as part of the Erlangen Vesicle Initiative. The project focuses on decoding microbial extracellular vesicle signatures in neuroinflammatory diseases by integrating high-dimensional omics data, including meta
genomics, proteomics, and metabolomics, from EVs isolated from stool and serum samples. By harmonizing and preprocessing these datasets, , as part of the Erlangen Vesicle Initiative. The project focuses on decoding microbial extracellular vesicle signatures in neuroinflammatory diseases by integrating high-dimensional omics data,
including metagenomics, proteomics, and metabolomics, from EVs isolated from stool and serum samples. By harmonizing and preprocessing these datasets, we apply machine learning models to identify robust, disease-stage–specific biomarkers and mechanistic fingerprints of relapse in multiple sclerosis.
To ensure the biological relevance and reproducibility of our findings, these models undergo rigorous validation through cross-cohort and cross-modality analyses. Through this work, our group advances the understanding of gut–brain communication via vesicle-based host–microbe interactions, offering broader implications for translational biomarker discovery in neuroimmunology. Ultimately, this integrative approach aims to uncover novel molecular signatures that can improve disease monitoring and therapeutic strategies in multiple sclerosis and related neuroinflammatory disorders.
Principal investigator: